DEGREES
2009 - Professor of Biological Sciences, nomination by the President of the Republic of Poland
2006 - DSc Habil, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznań, Poland
1999 - PhD in Biochemistry, Technical University of Munich, Germany
1995 - MSc in Experimental Physics, Munich University, Germany
PROFESSIONAL EMPLOYMENT
2011-present - Professor, Head of Laboratory of Structural in Biology, International Institute of Molecular and Cell Biology in Warsaw, Poland, and Laboratory of Genome Engineering, Institute of Biochemistry and Biophysics, Polish Academy of Sciences,Warsaw, Poland
2007-2011 - Part-time Director of Structural Biology, Cardiff University, United Kingdom
2001-2010 - Head, Joint MPG-PAS Junior Research Group, International Institute of Molecular and Cell Biology in Warsaw, Poland
2000 - Patent training, Weickmann & Weickmann
1999-2000 - Postdoctoral Fellow, Max Planck Institute of Biochemistry, Martinsried, Germany
RESEARCH TRAINING
1996-1999 - Research Assistant, Max Planck Institute of Biochemistry, Martinsried, Germany
1995-1996 - Internship, Medical Microbiology, University of Regensburg, Germany
1992-1993 - Guest Student, Cambridge University, United Kingdom
1990-1992 - Studies in Physics, Munich University, Germany
HONORS, PRIZES AND AWARDS
2018 - TEAM Foundation for Polish Science
2018 - International Academic Partnerships Programme, Polish National Agency for Academic Exchange
2018 - DAINA, National Science Centre
2015 - HARMONIA, National Science Centre
2012 - MAESTRO, National Science Centre
2011 - TEAM, Foundation for Polish Science Professor Stefan Pieńkowski Award
2004 - EMBO/HHMI Young Investigator Award
2000 - Crystal Award, Germany
1998 - Crystal Award Germany
1990 – 1992 - Scholarship from Deutsche Studienstiftung and Bavarian State
DOCTORATES DEFENDED UNDER LAB LEADER’S SUPERVISION
R. Filipek, M. Firczuk, M. Lipka, R. Szczepanowski, M. Kaus-Drobek, M. Sokołowska, G. Chojnowski, H. Korza, M. Wojciechowski, W. Siwek, P. Haniewicz, A.A. Kazrani, K. Mierzejewska.
The prevalent DNA modification in eukaryotes is C5-cytosine methylation. Biophysically, this modification stabilizes double-stranded DNA. Evolution has built on and enhanced this tendency to utilize C5-methylation, which predominantly occurs in the symmetric CpG context for transcription control. Consistent with its biophysical effects, methylation in transcription control structures (e.g., promoters) represses transcription; methylation elsewhere (e.g., gene bodies) enhances transcription by suppressing aberrant initiation. Methylation can be introduced in one step by de novo and maintenance methyltransferases and propagated by feed-forward loops that link DNA methylation and repressive chromatin marks. Methylation is most easily lost passively as a result of DNA replication, but it can also be actively erased through a mechanism that utilizes TET catalyzed oxidation to prime DNA for base excision repair. The genetics and cell biology of DNA methylation are unique to eukaryotes, but the biochemistry of DNA methylation (and to some extent also DNA demethylation) is also conserved in prokaryotes. We seek to answer biochemical questions using more robust bacterial proteins and answer genetic/ cell biological questions using zebrafish models and human genetic data (e.g., for malignancies with defects in demethylation machinery).
METHYLATION SENSING
In 2019, we continued our research on relatively well-behaved prokaryotic model proteins to study the specific recognition of 5-methylcytosine and its oxidized congeners in DNA. The repertoire of 5mC and 5hmC binding proteins is relatively small, and most proteins that specifically bind DNA with these bases contain the same domains. Domains that bind fully methylated DNA in the context of CpG include zinc finger and MBD domains. In contrast, hemi-methylated DNA is typically bound by SRA domains. During the last year, we considerably broadened the repertoire of known domains that recognize 5mC and 5hmC. SRA domains belong to the larger superfamily of PUA domains, which also comprises PUA domains in the strict sense, ASCH domains, EVE domains, and several other lesser known domain groups. To the extent that function was known, a clear division of labor appeared to be in place. SRA domains were associated with the binding of modified DNA, whereas other families within the PUA superfamily either were known to be involved in the binding or processing of modified RNA or had completely unknown functions. In silico screens that were performed in collaboration with Dr. Shuang-Yong Xu (New England Biolabs) showed that many PUA superfamily domains in bacteria are fused to endonucleolytic domains that are associated with DNA cleavage. Subsequent biochemical experiments demonstrated that the fusion proteins indeed cleaved modified DNA, although not with the same degree of specificity as SRA domains (Lutz et al., Nucleic Acids Res, 2019). We also crystallographically characterized a few prototype enzymes and solved their structures with and without DNA. The structures illustrate the mode of recognition of methyl- or hydroxymethyl modifications. Similar DNA modification-sensing domains exist in eukaryotes and have been implicated in malignancies, but their biochemical behavior requires further investigation. NEco is the modification-sensing domain of EcoKMcrA, which is one of the earliest studied restriction endonucleases of E. coli. EcoKMcrA efficiently restricts DNA that contains 5mC or 5hmC, provided the modifications are present in the right context. Efficiency is much greater for fully methylated DNA than for hemi-methylated DNA. Our previous work indicated that the NEco modification-sensing domain was phylogenetically unrelated to other methylationsensing domains. In 2019, we elucidated the binding mode of the domain to modified DNA and found that modification sensing is also locally very different from previous observations (Slyvka et al., Nucleic Acids Res, 2019). To date, NEco has been shown to be very good at discriminating (hydroxy)methylated from unmethylated DNA. Further research will likely expand the currently known and narrowly defined phylogenetic distribution (Fig. 1).
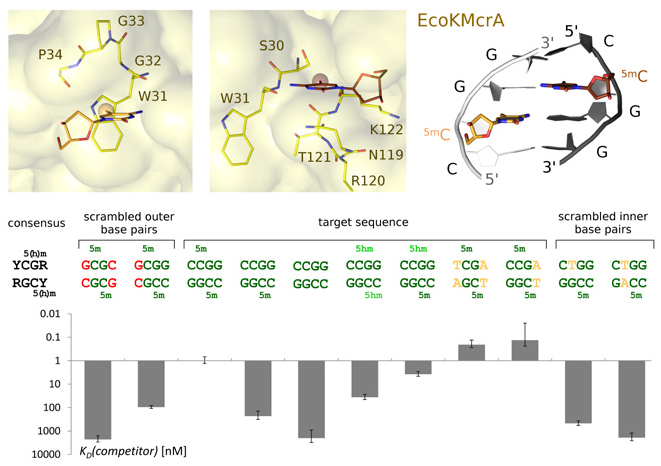
fig.1. Modification-dependent DNA binding by the N-terminal domain of EcoKMcrA endonuclease (NEco). (Top) Interaction between fully modified DNA with (hydroxy)methyl binding pockets of EcoKMcrA. (Bottom) Sequence and modification specificity of EcoKMcrA N-terminal domain determined by EMSA competition experiments (for experimental details, see Slyvka et al., Nucleic Acids Res, 2019).
DEMETHYLATION
In contrast to methylation sensing, demethylation has no clear equivalent in prokaryotes and thus needs to be studied using eukaryotic models. Our research primarily focuses on the ways in which TET proteins identify their targets. Two of our collaborators, Dr. Tomasz Jurkowski (Cardiff University, United Kingdom) and Dr. Tim Hore (Otago University, New Zealand), provided strong biochemical evidence that the locus specificity of TETs is at least partially attributable to sequence specificity. We solved structures with preferred and discriminated substrates to better understand the mode of sequence recognition. Based on the lack of sequence-specific contacts between TETs and their target DNAs outside the CpG core recognition sequence, we initially hypothesized that DNA bending was responsible for sequence specificity. Our crystal structures do not support this hypothesis, however, and instead reveal an unexpected mechanism of sequence recognition that also explains similarities in preference of different TET paralogues. We are also continuing our work on links between DNA reprogramming and DNA repair. Some of our experiments, such as investigating the role of NEIL1 and TDG in the excision of oxidized 5-methylcytosine bases, are consistent with the general paradigm that DNA reprogramming co-opts DNA repair. However, based on much circumstantial evidence and the work of others, we suspect that the converse may also be true and that DNA repair may use intermediates that are normally associated with reprogramming. We will test this hypothesis both biochemically and bioinformatically.

Lab Leader:
Senior Researcher:
Postdoctoral Researchers:
PhD Students:
Another co-worker:
Lab Technician:
Laboratory Support Specialist:
Contact:
This email address is being protected from spambots. You need JavaScript enabled to view it.



