 |
Wojciech Pokrzywa, PhD, DSc Habil
Correspondence address:
Laboratory of Protein Metabolism
International Institute of Molecular and Cell Biology
4 Ks. Trojdena Street, 02-109 Warsaw, Poland
Email: This email address is being protected from spambots. You need JavaScript enabled to view it.
tel: +48 (22) 597 0742; fax: +48 (22) 597 0715
|
DEGREES:
2020 - DSc Habil in Biological Sciences, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Poland
2009 - PhD in Biological Engineering and Agronomic Sciences at the Institute of Life Sciences, Molecular Physiology Group (FYMO), Catholic University of Louvain, Belgium.
2006 - Master of Advanced Science in Biological Engineering and Agronomic Sciences at the Catholic University of Louvain, Belgium.
2004 - Master’s in Microbiology at the University of Wroclaw, Poland.
PROFESSIONAL EXPERIENCE
2017-present - Professor, Head of Laboratory of Protein Metabolism in Development and Aging, International Institute of Molecular and Cell Biology in Warsaw, Poland
2009-2017 - Postdoctoral fellow at the Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Germany.
2004-2008 - PhD studies at the Institute of Life Sciences, Molecular Physiology Group (FYMO), Catholic University of Louvain, Belgium.
HONORS, PRIZES AND AWARDS
2018 - FIRST TEAM, Foundation for Polish Science
2018 - EMBO Installation Grant
2017 - OPUS, National Science Centre
2005 - PhD Fellowship from the FNRS-Fund for Scientific Research, Belgium
2004 - ERASMUS Scholarship
DESCRIPTION OF CURRENT RESEARCH
The proteome is defined as the entire set of proteins that are expressed in a given cell type or organism, which can vary with time and physiological status. Quality control networks support the integrity of the cellular proteome. The human protein homeostasis network (proteostasis) involves >1000 accessory factors and regulatory components that govern protein synthesis, folding, and degradation. Defective folding can result in a greater abundance of toxic protein aggregates, which can endanger the integrity of the entire proteome. With age, the ability of post-mitotic cells to maintain a stable proteome is gradually compromised, particularly by the downregulation of molecular chaperones and lower efficiency of protein degradation. As such, impairments in proteostasis are a major hallmark of aging and associated with dementia, neurodegeneration, type 2 diabetes, cystic fibrosis, cancer, and cardiovascular disease (Labbadia and Morimoto, Annu Rev Biochem, 2015). One of the central nodes of the eukaryotic proteostasis network is the interaction between molecular chaperones and proteolytic machinery. To maintain the cellular proteome, molecular chaperones and ubiquitin-dependent degradation pathways coordinate protein refolding and the removal of terminally damaged proteins. Irreversibly affected proteins are recognized by chaperone-assisted E3 ubiquitin ligases, which target them for degradation by the ubiquitin-proteasome system (UPS) or autophagy (Fig. 1). Our studies concentrate on the basic understanding of the spatiotemporal regulation of protein quality control activity and processing of its substrates. In our research, we use a combination of biochemical, microscopic, and molecular genetic techniques and tissue-specific approaches in C. elegans.
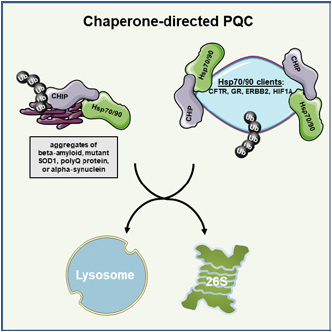
fig.1. The illustration represents an important component of the protein homeostasis system, the quality control ubiquitin ligase CHIP. The primary function of CHIP is polyubiquitylat ion of chaperone-bound polypeptides facilitating the switch from chaperone-mediated folding/maturation to proteasomal degradation. In cooperation with chaperones, CHIP ameliorates proteotoxicity in various proteinopathies by marking aggregates of beta‐amyloid, mutant SOD1, or alpha‐synuclein for degradation. In addition to misfolded proteins, CHIP promotes degradation of a broad array of substrates when bound to chaperones, such as glucocorticoid receptor or hypoxia-inducible factor 1. Figure from Kevei, Pokrzywa, Hoppe. FEBS Lett, 2017
WE FOCUS MAINLY ON THE FOLLOWING PROJECTS:
The fate of eukaryotic proteins, from their synthesis to destruction, is supervised by the UPS. The UPS is the primary pathway that is responsible for the selective proteolysis of intracellular proteins, which is guided by the covalent attachment of ubiquitin to target proteins by E1 (activating), E2 (conjugating), and E3 (ligating) enzymes in a ubiquitylation process. Despite many structurally unrelated substrates, ubiquitin conjugation is remarkably selective. E3 ubiquitin ligases represent the largest group of proteins within the UPS, which is linked to their crucial role in substrate selection. A detailed analysis of several classes of E3 ligases identified specific proteins and molecular pathways that they regulate.
Furthermore, the heterotypic oligomerization of E3 ligases might control the specificity and processivity of ubiquitylation. Scott et al., (Cell, 2016) reported that two distinctive E3s could reciprocally monitor each other for the simultaneous and joint regulation of substrate ubiquitylation. Cullin-RING (CRL) ligase was shown to associate with a mechanistically distinct thioester-forming RBR-type E3, ARIH1, and rely on ARIH1 to directly add ubiquitin chains on CRL substrates.
A combination of E3 ligases could support the formation of alternative ubiquitylation structures in different physiological processes, which probably allows them to improve substrate recognition and ubiquitylation process. Our long-term objective is to understand the mechanistic and developmental aspects of protein degradation pathways that are defined by a specific pair of E3 enzymes.
S-adenosyl-L-methionine (SAM)-dependent methylation is central to the regulation of many biological processes, including gene expression, signaling, protein synthesis, and lipid metabolism. SAM is synthesized in the cytosol of every cell from L-methionine and adenosine triphosphate in a reaction that is catalyzed by methionine adenosyltransferase (Fontecave et al., Trends Biochem, 2004). Despite fundamental roles of the SAM cycle in a broad range of biological processes, the mechanisms of its regulation are still enigmatic. Our preliminary studies suggest that the UPS regulates SAM cycle activity. We aim to understand the ways in which the UPS modulates methionine metabolism, methylation potential of the cell, and epigenetic memory using C. elegans as an animal model. Our research may have broad implications for understanding the regulation of methionine metabolism in both health and disease.
The assembly and maintenance of myofilaments require a tightly balanced proteostasis network. One key player in myosin organization and muscle thick-filament formation in health and disease is the Hsp90 co-chaperone UNC-45. The activity and assembly of various myosin subtypes are coordinated by conserved UCS (UNC-45/CRO1/ She4p) domain proteins. One founding member of the UCS family is Caenorhabditis elegans UNC45, a protein that is essential for the organization of striated muscle filaments (Price et al., J Cell Sci, 2002). Moreover, UNC-45 homologs exist in vertebrates, indicating a conserved requirement for myosin-specific co-chaperones. Indeed, abnormal UNC-45 function is associated with severe muscle defects that result in skeletal and cardiac myopathies (Janiesch et al., Nat Cell Biol, 2007).
The integrity of sarcomeric structures is permanently challenged upon muscle growth and mechanical stress. In response to eccentric exercise or damage to myofibers, UNC-45 and the chaperone Hsp90 shuttle between the impaired myofibers to support their repair (Fig. 2). However, little is known about the coordination of protein homeostasis pathways upon mechanical stress. Therefore, the long-term objective of this project is to understand the ways in which the balance between protein folding and degradation networks is coordinated with myosin assembly and muscle integrity. We combine genetic and biochemical approaches to study the conserved function of UNC-45 in myosin assembly and examine the ways in which this function is modulated during mechanical stress. Specifically, we plan to use targeted screening strategies to uncover mechanosensory proteins, chaperones, and UPS and autophagy components that are required for muscle function. The conserved regulation of proteostasis networks is studied in C. elegans, C2C12 mouse myoblasts, and human skeletal muscles. Finally, we are investigating the remodeling of UNC-45 folding machinery under conditions of mechanical stress.
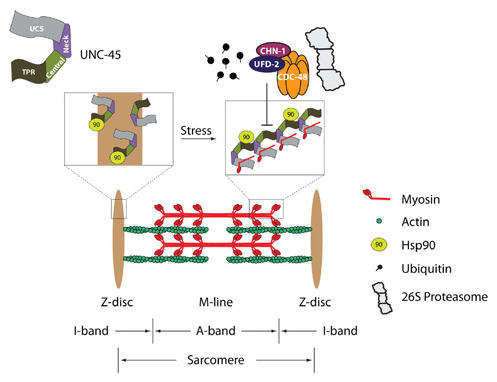
fig.2. Model for the UNC-45 polymerization in response to stress. The sarcomeric unit is defined by the distance between two Z-discs including the A-band, I-band, and M-line. UNC-45 composes tandem modules that allow the simultaneous binding of Hsp70/Hsp90 and myosin, enabling the folding and assembly of myosin in regular spacing. In the fully developed muscle, monomeric UNC-45 might be stored at the Z-disk, which anchors the thin actin filaments of the I-band. Under stress conditions, UNC-45 is relocated to damaged myosin filaments of the A-band and might assemble into short chaperone chains to maintain the sarcomeric structure especially during muscle regeneration and aging. The conserved CDC-48/UFD-2/CHN-1 ubiquitylation complex might influence the process of UNC-45 chain formation. The ubiquitylation of UNC-45 either reduces the pool of the monomeric form available for chain formation or inhibits UNC-45 polymerization directly by modifying the binding interface. Figure adapted from Pokrzywa and Hoppe, Worm, 2013.

Lab Leader:
Postdoctoral Researchers:
Researcher:
Research Technician:
PhD Students:
-
Katarzyna Banasiak, MSc
-
Aniruddha Das, MSc
-
Pratik Kumar Mandal, MSc
-
Pankaj Thapa, MSc
-
Anwesha Sarkar, MSc
-
Natalia Szulc, MSc
Trainees:
-
Marta Jaśkiewicz
-
Konrad Kowalski
Lab Technician:
Laboratory Support Specialist:




