 |
Jan Brezovsky, PhD |
DEGREES
2011 - PhD in Environmental Chemistry, Faculty of Science, Masaryk University, Brno, Czech Republic
2007 - MSc in Biophysics, Faculty of Science, Masaryk University, Brno, Czech Republic
PROFESSIONAL EXPERIENCE
2016 - present - Professor, Head of the joint laboratory, International Institute of Molecular and Cell Biology in Warsaw and Institute of Molecular Biology and Biotechnology, Faculty of Biology, Adam Mickiewicz University in Poznań, Poland
2016 - Assistant Professor, Department of Experimental Biology, Masaryk University, Brno, Czech Republic
2015-2016 - Postdoctoral Researcher, International Clinical Research Center, St. Anne’s University Hospital, Brno, Czech Republic
2014 - Research visit to the group of Prof. Rebecca Wade, Heidelberg Institute of Theoretical Science, Germany
2012-2016 - Leader of Research Team, Loschmidt Laboratories, Faculty of Science, Masaryk University, Czech Republic
2009 - 2011 - Research Assistant, Loschmidt Laboratories, Department of Experimental Biology, Faculty of Science, Masaryk University, Brno, Czech Republic
2007 - 2008 - Research Assistant, National Centre for Biomolecular Research, Faculty of Science, Masaryk University, Brno, Czech Republic
HONORS, PRIZES AND AWARDS
2018 - SONATA BIS, National Science Centre
2017 - OPUS, National Science Centre
2016 - GACR grant, Czech Science Foundation
2015 - 2016 - Elected member of the national node committee of European Life-Science Infrastructure for Biological Information, Czech Republic (ELIXIR-CZ)
2011 - 5th place at national competition Chemistry Prize of Jean-Marie Lehnen
2011 - Dean’s prize for outstanding PhD research, Masaryk University, Brno, Czech Republic
2007 - Research grant from Masaryk University, Brno, Czech Republic
Research in our laboratory is oriented toward answering fundamental questions about the mechanism of action of various proteins that have biomedical and biotechnological importance. We investigate the mechanisms that enable the migration of ligands to and from functional sites that are deeply buried within protein structures. We also explore the implications of such processes for the functions of living cells. To achieve these goals, we develop new computational protocols and tools and apply them to the analysis of biomedically and biotechnologically relevant proteins.
At any given moment, living systems contain several thousand small organic molecules, both endogenous and exogenous, comprising the metabolome. To exert their function, the hosts of molecules need to arrive at their sites of action, mostly represented by protein surfaces and internal cavities. The transport of the metabolome is largely governed by protein tunnels and channels. Such tunnels and channels secure the transport of ligands between different regions and connect inner protein cavities with the protein surface, connect two or more different cavities, or connect even different cellular environments, such as in membrane proteins. The presence of very sophisticated transport processes markedly contributes to the symbiotic co-existence of individual chemical species within a single compartment or whole cell without the presence of overly disruptive interference. Protein channels facilitate the regulated and very selective transport of ions and ligands across a membrane between different cellular compartments. The role of channels in the function of various proteins has been the focus of intense research for years. Their importance is illustrated by many diseases that are caused by channel mutations. Such channel pathologies can severely impair the function of many physiological systems, manifested as various diseases, including epilepsy, hypertension, cystic fibrosis, diabetes, and cancer. To counteract these malfunctions, many inhibitors or activators that affect transport through these channels have been identified.
Tunnels connect buried functional sites to the bulk solvent, enabling the access of substrates and release of products. Moreover, the tunnels are responsible for many additional functions that are essential for the proper actions of proteins that are exposed to interference from individual species that are present in the metabolome of the living cell. The tunnels enable the access of preferred substrates and deny access to non-preferred substrates. The tunnels can prevent damage to enzymes that contain transition metals through ligation and damage to the cell that is caused by the release of toxic intermediates to the cellular environment. The tunnels also enable reactions that require the absence of water and allow the temporal and spatial synchronization of reactions. Most enzymes likely possess tunnels. In fact, the presence of tunnels was already described for enzymes from six Enzyme Commission classes and four structural classes of proteins. In many cases, tunnels are transient, meaning they cannot be readily identified from static crystal structures. Therefore, we can expect the discovery of tunnels in many other protein families. Recognizing the importance of transport processes for enzymatic catalysis, many protein engineering studies have successfully modified tunnels to improve enzymatic activity, specificity, enantioselectivity, and stability. Tunnels were established as critical functional factors in enzyme catalysis relatively recently, and their role in cellular biochemistry and tunnel mutations in disease etiology has been largely overlooked. However, many enzymes that are known to contain tunnels have been associated with the development of various ailments, including cancer, neurodegenerative disorders, autoimmune diseases, and inflammation. Inhibitors of some of these enzymes have been shown to bind to tunnels exclusively, thus confirming the proposed role of tunnels in disease etiology and treatment.
TO FILL THE GAPS IN OUR KNOWLEDGE OF LIGAND TRANSPORT PHENOMENA, WE ARE CURRENTLY FOCUSING ON THE FOLLOWING:
Enabling routine and reliable analysis of transient transport tunnels in proteins
The primary goal of this project is to enable large-scale studies of properties and dynamics of functionally relevant transport tunnels. We are currently evaluating and optimizing various approximate dynamics methods to provide ensembles of protein structures with tunnel properties and dynamics that correspond to those that are obtained from rigorous simulations. In the next step, we will utilize the developed and thoroughly validated method for the detection of transient tunnels in all proteins with buried functional cavities with available 3D structures. The biologically relevant tunnels that are identified in these proteins will extend our fundamental knowledge about the properties that determine the transport component of protein function. We believe that the results of this large-scale analysis will stimulate further investigations of transient tunnels and their gates and enable the targeting of transient tunnels in protein engineering and drug discovery efforts (Fig. 1).
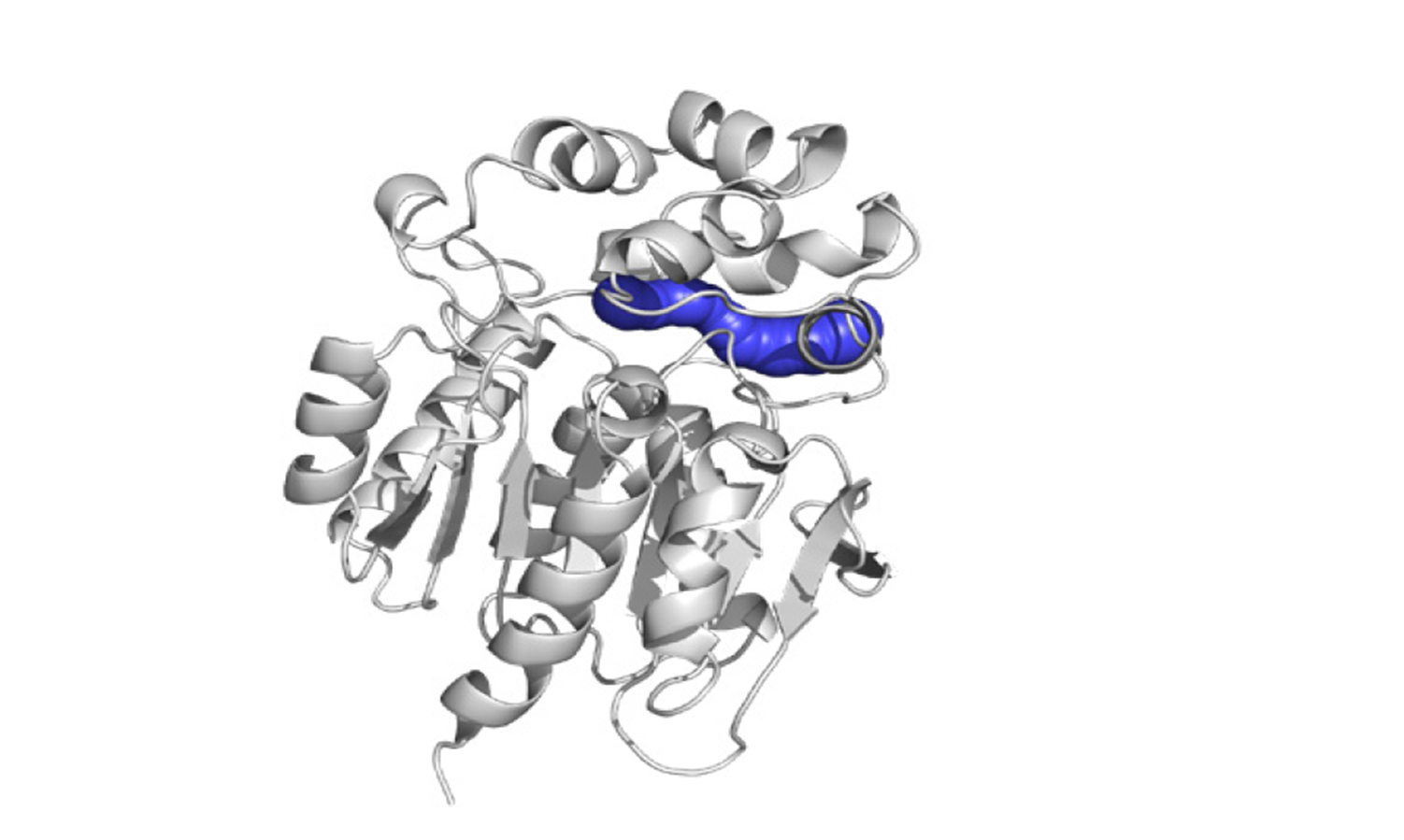
fig.1. A prospective transient tunnel (blue spheres) that forms a “back-door” to the active site of haloalkane dehalogenase DhaA (white cartoon) as identified from analyses of massive molecular dynamics simulations, capturing a total of 5 μs of simulation time.
Understanding molecular origins of mechanisms that govern functions of enzymes with buried actives sites
The primary goal of this research is to unveil molecular bases of largely unexplored factors that notably affect the biological function of enzymes with buried active sites (i.e., substrate inhibition, cooperativity, and interference between molecules of substrates and products during their simultaneous transport via the tunnels). We are working on comprehensive kinetics models of ligand transport in enzymes that will enable us to perform detailed analyses of structure-dynamics-function relationships that govern the transport of multiple ligands via the tunnels, revealing roles of direct interactions among ligands and allosteric effects on the tunnels that are mediated by protein-ligand interactions. We expect to obtain novel insights into the origins of substrate inhibition and cooperativity (i.e., phenomena that are necessary for the proper in vivo functions of many enzymes). The knowledge that is obtained can then be exploited to target these properties in research that seeks to engineer better enzymes and develop novel inhibitors. Finally, the findings on mutual interference among different ligands and effects on their transport will facilitate more accurate studies of enzyme-drug association/dissociation processes, thus paving the way toward the optimization of drug residence times.

Lab Leader
Jan Brezovsky, PhD
Postdoctoral Researchers
Nikhil Agrawal, PhD (AMU)
Cedrix Dongmo Foumthuim, PhD (until November 2019)
PhD Students
Nishita Mandal, MSc (AMU)
Dheeraj Kumar Sarkar, MSc (AMU)
Carlos Eduardo Sequeiros Borja, MSc
Bartłomiej Surpeta, MSc
Aravind Selvaram Thirunavukkarasu, MSc
Undergraduate Students
Wiktor Szymkowski, BSc (AMU)
Research Technician & Laboratory Support Specialist
Katarzyna Voelkel, MSc



