 |
Gracjan Michlewski, PhD, Professor
Correspondence address:
Laboratory of RNA-Protein Interactions - Dioscuri Centre
International Institute of Molecular and Cell Biology
4 Ks. Trojdena Street, 02-109 Warsaw, Poland
E-mail: This email address is being protected from spambots. You need JavaScript enabled to view it.
|


|
DEGREES
2021 - Professor of Biological Sciences, nomination by the President of the Republic of Poland
2012 - DSc Habil in Biochemistry, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznań, Poland
2005 - PhD in Biological Chemistry summa cum laude, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznań, Poland
2001 - MSc in Biotechnology, Adam Mickiewicz University, Poznań, Poland
PROFESSIONAL EXPERIENCE
2021-present - Professor, Head of Laboratory of RNA-Protein Interactions - Dioscuri Centre
2021-present - Editorial Board Member, Communications Biology, Nature Group
2021-present - Honorary Lecturer, Infection Medicine, The University of Edinburgh, Edinburgh, United Kingdom
2020 - Reader, Infection Medicine, The University of Edinburgh, United Kingdom
2018-2020 - Associate Professor, Zhejiang University-University of Edinburgh Institute, Haining, China
2018-2020 - Senior Lecturer, Infection Medicine, The University of Edinburgh, Edinburgh, United Kingdom
2011-2017 - Medical Career Award Fellow, Wellcome Trust Centre for Cell Biology, The University of Edinburgh, Edinburgh, United Kingdom
2005-2010 - Post-doctoral fellow, Human Genetics Unit, Medical Research Council, Edinburgh, United Kingdom
HONORS, PRIZES, AND AWARDS
2021-2025 - Polish Returns Programme, Polish National Agency for Academic Exchange
2021-2025 - Dioscuri Centre for RNA-Protein Interactions in Human Health and Disease, The Max Planck Society and The National Science Centre Poland
2020 - AIMS Award, Atomwise
2019-2022 - Project Grant, UK Government’s Biotechnology and Biological Sciences Research Council2018 Award, Moray Endowment Fund
2017-2019 - Seed Award in Science, Wellcome Trust
2017 - Travel Grant, RNA Society
2011 - 2015 Career Development Award, Medical Research Council
2010 - Scholarship, Keystone Symposia
2010 - International Travel Grant, The Royal Society
2008 - Scholarship, Keystone Symposia
2004-2006 - Award for Scientific Achievements, Polish Genetic Society
2001 - Fellowship Award, Minister of Polish National Education
2001 - Fellowship Award, Adam Mickiewicz University Foundation
DOCTORATES DEFENDED UNDER LAB LEADER’S SUPERVISION
J.S. Nowak, B. Özkan, G. Heikel, A. Downie Ruiz Velasco
See more: Dioscuri Centre for RNA-Protein Interactions in Human Health and Disease
Dioscuri Centre of Scientific Excellence. The Programme initiated by the Max Planck Society (MPG), managed jointly with the National Science Centre in Poland and mutually funded by the Polish Ministry of Education and Science (MEiN) and the German Federal Ministry of Education and Research (BMBF).

RNA-PROTEIN INTERACTIONS IN HUMAN HEALTH AND DISEASE
RNA-binding proteins (RBPs) are key molecules that control gene expression signalling through RNA-protein interactions. Consequently, they contribute to cellular homeostasis, normal development and majority of human diseases. Importantly, new RBPs are being discovered by high throughput proteomics, but we still have a limited understanding of their function. A pioneering study identified 860 RNA-binding proteins in HeLa cells (Castello et al., Cell 2012). Strikingly, approximately 300 of these proteins were not known for their RNA-binding properties and bear no identifiable RNA-binding domains. Several other studies identified hundreds of novel RNA-binding proteins in various cell and tissues. Despite the extensive cataloguing of novel RNA-binding proteins, we have a limited understanding of what dictates their molecular function and of their RNA-related roles in human health and disease.
We are currently focusing on two main topics:
-
Functional and structural characteristics of novel RBPs and RNA-protein interactions in innate immune response to RNA viruses
-
Regulation of microRNAs through RBPs for the treatment of Parkinson’s disease
HOST-VIRUS RNA-PROTEIN INTERACTIONS
RNA viruses have caused several epidemics in the 21st century. Currently raging SARS-CoV-2 virus, causing COVID-19, killed already 2.2 mln people and wiped $10 trillion from the global economy. Another well-known RNA virus - IAV (influenza A virus) kills up to 500,000 people annually and generates a significant global socioeconomic burden. These are only two examples of many RNA viruses that cause severe problems for human health and economy. Thus, a detailed molecular understanding of host-virus interactions is imperative in order to know how best to inactivate the virus and prevent major disruptions. Finally, viruses have been used to uncover some of the most important cellular processes such as mRNA splicing, capping, polyadenylation or RNA interference and they continue providing insights into molecular phenomena that can aid in understanding basic biology of living organisms.
We have recently discovered and started characterising novel RNA binding protein – E3 ubiquitin ligase TRIM25 (Choudhury et al., Cell Reports 2014; Choudhury et al., BMC Biology 2017). TRIM25 belongs to a large family of tripartite motif-containing proteins (more than 80), most of which have E3 ubiquitin ligase activity. All TRIMs have in common their amino-terminal tripartite domain arrangement - RING–Bbox1/2–coiled coil (CC) - yet differ in their C-terminal domains, which categorize them into a number of subtypes. Many of TIRIMs are positive or negative regulators of innate immune pathways – a first line of defence against pathogens such as viruses. Importantly, TRIM25 is emerging as a key factor in the innate immune response to RNA viruses (including IAV, SARS, sindbis virus (SINV), dengue virus and many others). Despite the essential involvement of TRIM25 in viral RNA-induced innate immune pathways, its RNA-binding functions are still poorly understood.
The most widely reported TRIM25 function is in activating the pattern-recognition receptor RIG-I that senses 5’-triphosphate (5’-ppp) moieties on viral RNAs and activates innate immune response. Upon binding to 5’-ppp-RNA, RIG-I undergoes TRIM25-mediated K63 ubiquitination. This activates a signalling cascade that culminates in the phosphorylation of IRF-3, IRF-7 and NF-κB, which translocate to the nucleus and induce type I interferon expression. Importantly, TRIM25’s role in RIG-I pathway has been recently challenged by numerous reports. We have discovered that TRIM25 is an RNA-binding protein that regulates the stability of host RNA (Choudhury et al., Cell Reports 2014). We have also revealed that TRIM25 binds RNA through a novel RNA-binding domain (residing in the PRY/SPRY region) and showed that RNA binding appears to be crucial for its E3 ubiquitin ligase activity (Choudhury et al., BMC Biology 2017) (Fig. 1). Despite this, direct binding of TRIM25 to viral RNAs, its 3D structure in the complex with host or viral RNAs and its detailed function in cell biology and innate immunity have not yet been described. Also, the RNA-binding potential of other TRIMs with PRY/SPRY domains have not yet been explored.
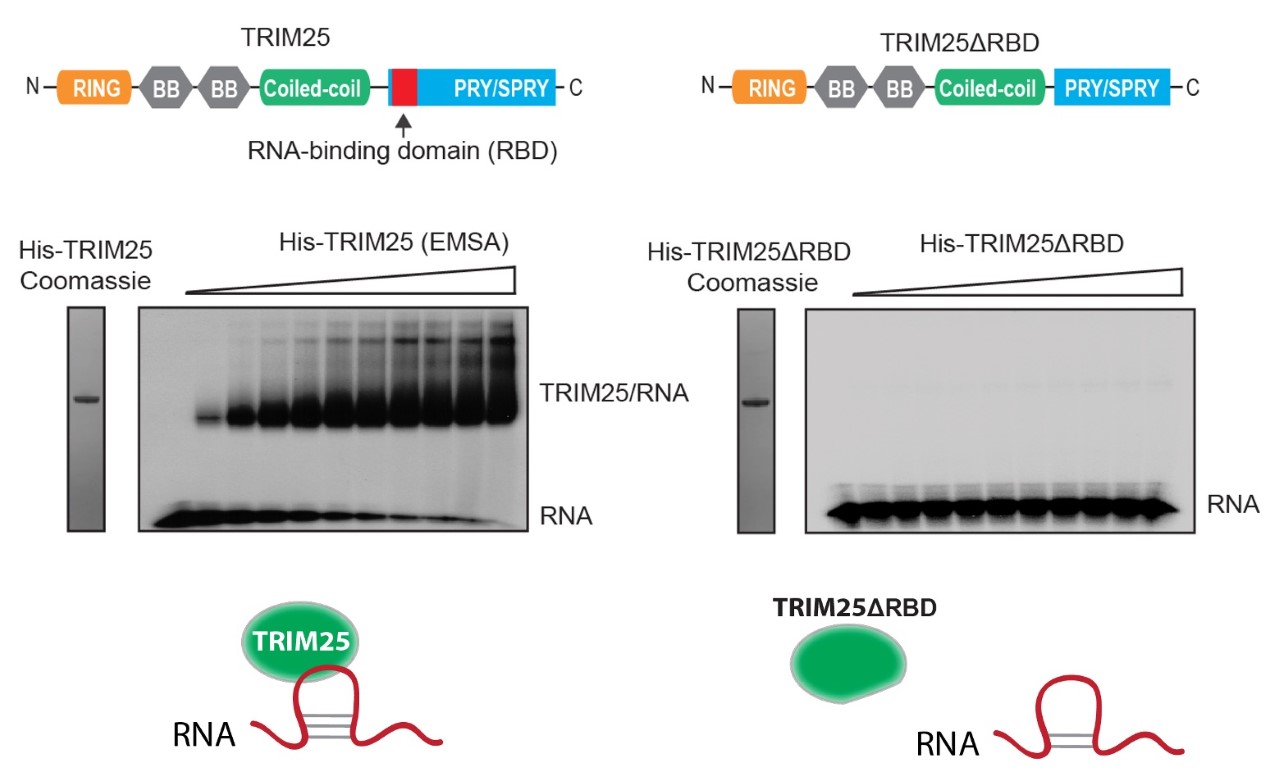
Fig. 1. The E3 ubiquitin ligase TRIM25, a key factor in innate immune response to RNA viruses is an RNA-binding protein. TRIM25 binds RNA through its PRY/SPRY domain (Choudhury et al., BMC Biology 2017).
Within the same theme, we have recently co-authored a study that identified global changes in RBPs composition and affinities in response to SINV infection (Garcia-Moreno et al., Molecular Cell 2019). Over two hundred RBPs showed differential association with RNA upon infection with SINV. This remodelling of RNA-protein interactions is driven by the host cell to limit virus infection and by virus to limit host innate immune response.
We are currently working towards uncovering the roles of the novel RNA-protein interactions in the innate immune response to RNA virus infections. Concomitantly, we aim to solve the E3 ubiquitin ligase TRIM25/RNA complex structure, which will inform us about the novel RNA-binding domain(s) and the role of RNA in stimulating ubiquitination. The outcome of this research will reveal how TRIM25 uses its RNA-binding activity for anti-viral functions. Importantly, TRIM25 belongs to a large (> 80 members) family of tripartite motif-containing proteins. They have various functions in cellular processes and disease, including development, apoptosis, autophagy, carcinogenesis and innate immunity. Thus, our research will open new lines of investigations into other TRIM proteins and their putative RNA-binding roles.
REGULATION OF MICRORNA BIOGENESIS
MicroRNAs (miRs) are small noncoding RNAs that negatively regulate the expression of mRNAs. They have defined tissue expression patterns and affect many cellular processes and developmental pathways. Most miRs are transcribed by RNA polymerase II, with the long primary transcript, termed pri-miR, carrying a hairpin structure. The biogenesis of miRs is carried out by two enzymes, DROSHA and DICER, which catalyse two processing events, in the nucleus (from pri-miR to pre-miR) and in the cytoplasm (from pre-miR to miR duplex), respectively. MiR duplexes are incorporated into the RISC (RNA-induced silencing complex) together with an Argonaute (AGO) protein, where one strand is selected to become the mature miR. The RISC then recognizes a specific mRNA sequence by complementary base-pairing resulting in translation inhibition and/or RNA degradation.
Due to the important role of miRNAs in the control of gene expression and organism development, the production of mature miRNAs is tightly regulated at multiple levels, including transcriptional and post-transcriptional steps. We have shown that the biogenesis of miRs can be regulated post-transcriptionally by RBPs that binds to their pri-miRs and pre-miRs (Michlewski et al., Mol Cell 2008, Michlewski and Caceres Nat Struct Mol Biol 2010, Choudhury et al., Genes Dev 2012, Nowak et al., Nat Commun 2014, Nowak et al., RNA 2017, Kooshapur et al., Nat Commun 2018, Michlewski and Caceres RNA 2019, Downie Ruiz Velasco et al. Mol Ther Nucleic Acids 2019, Sajini et al. Nat Commun 2019).
Currently we are working on regulation of miR-7 pathway, which is directly involved in Parkinson’s disease (PD). PD is an incurable neurodegenerative disease that affects all ages but is most prevalent in elderly with over 1% of the population over the age of 60 suffering from this disease. At present, there are about 10 million people living with PD and hundreds of thousands die each year from this devastating disease. One of the main causes behind PD is overproduction and aggregation of a protein called alpha-synuclein (alpha-Syn) in the brain cells of affected individuals. There is a large body of evidence that decreasing the levels of alpha-Syn should be beneficial for PD patients and several clinical trials are now focusing on alpha-Syn clearance with silencing RNAs or vaccines. Notably, miR-7 has been shown to target alpha-Syn production and approaches for miR-7 replacement therapies have been put forward.
We have shown that HuR (ELAVL1) protein is a naturally occurring inhibitor of miR-7 production that binds to its pri-miR-7 CTL (Choudhury et al., Genes Dev 2012). Furthermore, evidence suggests that HuR is upregulated in PD and that binding to alpha-Syn mRNA 3’UTR is stabilizing the transcript, allowing increased alpha-Syn production. This suggests that disrupting the RNA/HuR complex will have a positive effect on miR-7 and negative effect on alpha-Syn, providing novel approach for PD therapy.
In the last few years, we have been developing novel drug discovery platform, based on RNA-Protein Interactions in Eukaryotic Cell Extracts with Confocal Nanoscanning (RP-CONA) to screen for specific compounds interfering with the pri-miR-7/HuR complex, thus reactivating miR-7 biogenesis, suppressing alpha-Syn production and alleviating PD symptoms (Zhu et al., bioRxiv 2021). Our strategy is based on the use of an ultra-sensitive RNA-protein interaction assay in cell extracts from human cultured cells (Choudhury and Michlewski et al, Methods 2019) and detecting pri-miR-7/HuR complex disruption by confocal nanoscanner (Fig. 2). The use of human cultured cell extracts for an ultrasensitive on-bead assay offers great advantages over traditional systems for drug screening that use purified proteins from prokaryotic cells. Firstly, all human proteins, such as HuR are extensively modified in human cells, which directly influence their structure, activity and function. Secondly, RNA-protein complexes, such as pri-miR-7/HuR are analysed in the context of other competing pri-miR-7/protein complexes, better reflecting cellular environment. Our new methodology will allow discovery of compounds that disrupt pri-miR-7/HuR complex, as close to the human cellular context as possible. Finally, the implementation of RP-CONA screening platform will allow many researchers to explore the possibilities of targeting RNA-protein interactions in research and medical-based projects.
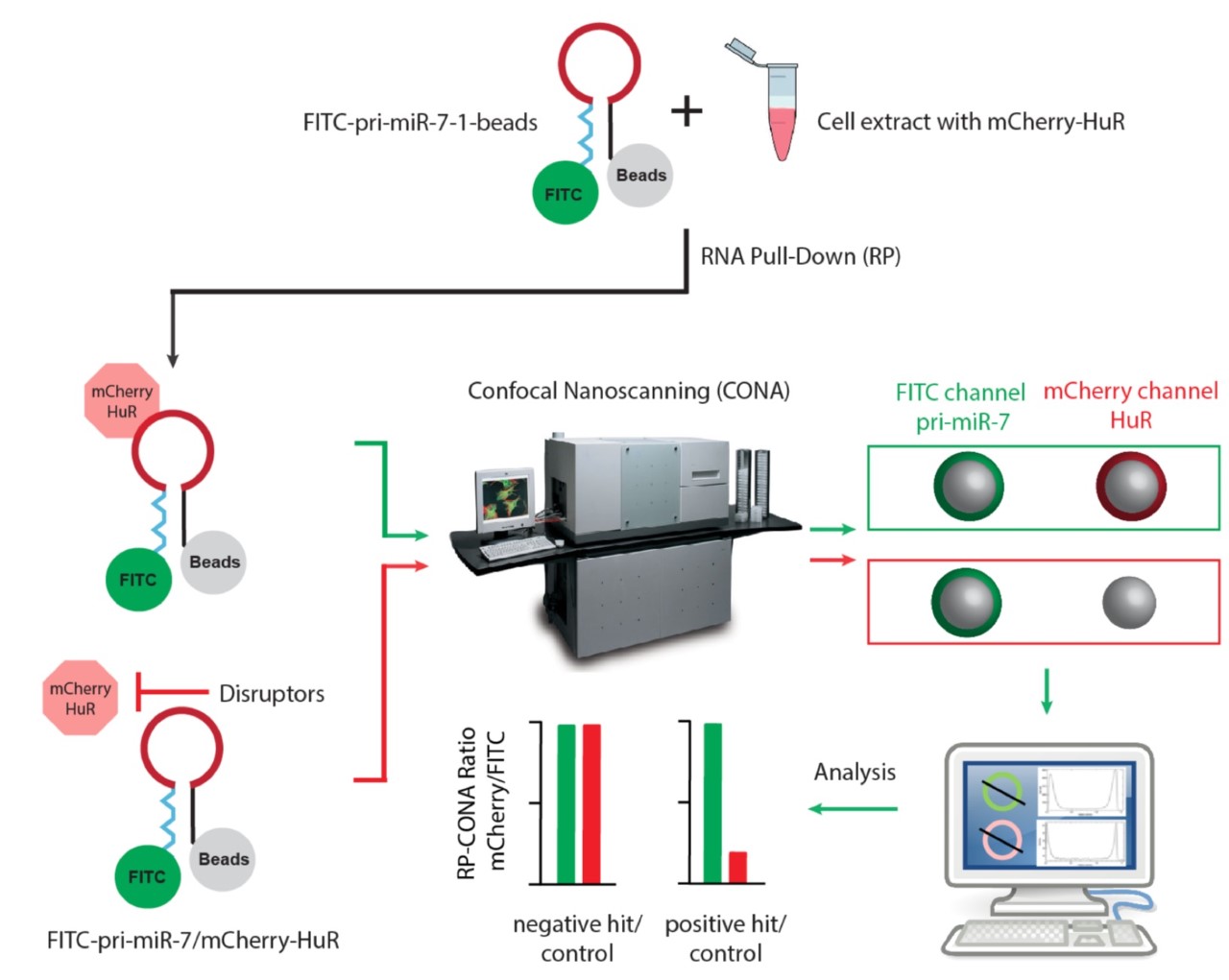
Fig. 2. The working scheme of RP-CONA, a novel drug discovery platform for RNA-protein interaction (Zhu et al., bioRxiv 2021).
Lab Leader
Gracjan Michlewski, PhD, Professor
Postdoctoral Researchers
Ivan Trus, PhD
Magdalena Wołczyk, PhD
Research Assistant
Natalia Stec, MSc
PhD Student
Agnieszka Bolembach, Msc
Other Co-worker
Siran Zhu, MSc
Laboratory Support Specialist
Monika Myśliwiec, MSc






